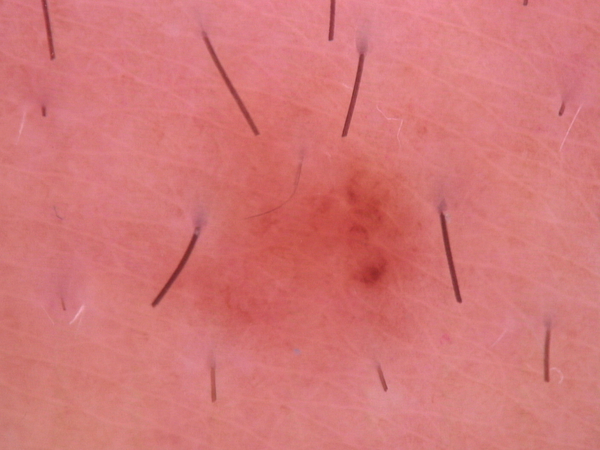
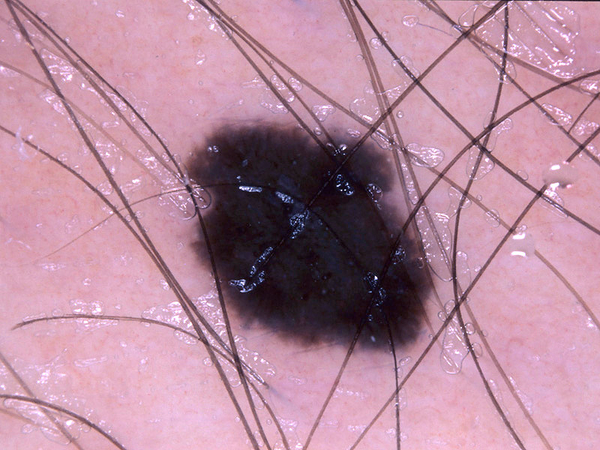
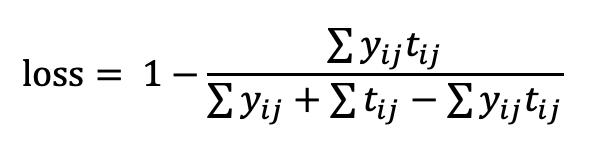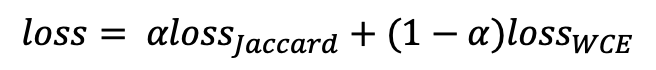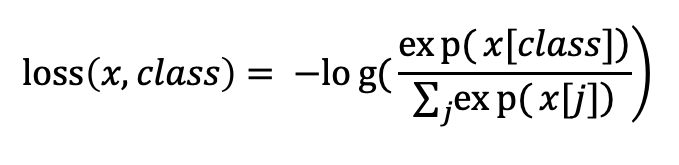Image Pre-processing
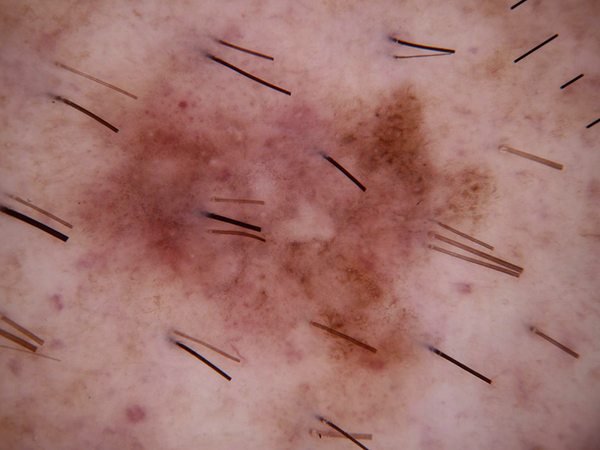
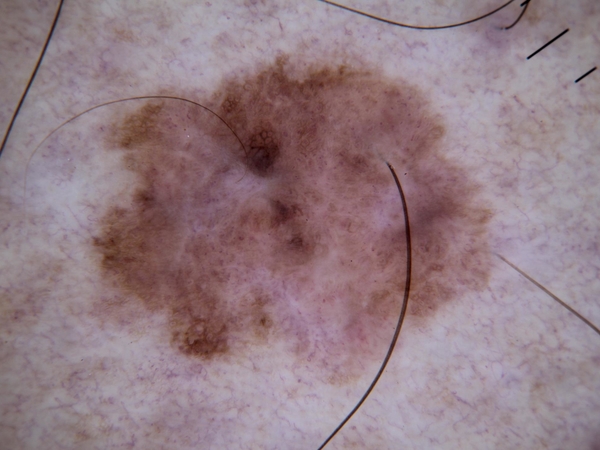
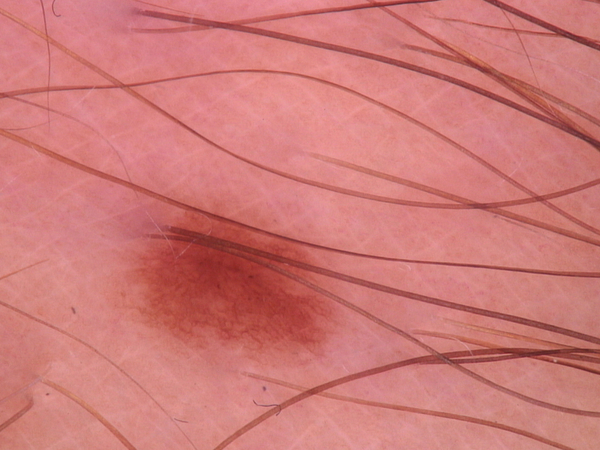
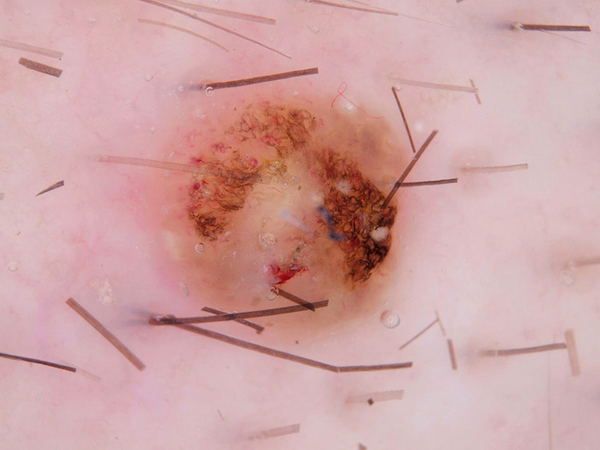
By observing the dermoscopic images, we found out the characteristic of hair noises include: they normally looks slender and not circle, and they normally is darker than surrounding skin. So my detection method is based on these two points.
Adaptive Thresholding
First, I found out the areas which are relatively darker than neighbors by adaptive thresholding. Adaptive thresholding is an extend in thresholding: in normal thresholding, the reference threshold is based on the mean of whole range; in adaptive thresholding, the threshold for each point is the mean calculated based on the neighbor area. The advantage of adaptive thresholding includes dynamic block size and threshold offset.
Geometric Shape
After filtered out all the related-darker area, the next step is to pick out the hair area in these contours. Under the first selection, hair, scar and stage micrometer will be collected. Hair and stage micrometer are noise to a disease image, so both of them are chosen to remove from images. There are some difference in the two group of characteristic which I want to separate: for hair and stage micrometer, they usually look slender; for scar, it usually has a hollowed look and might cross large area. I will separate them based on their geometric shape. I used 2 methods to calculated their difference:
1. Ratio between radius of external circle(r) and perimeter(s):
- s/r = 2𝜋 : this contour is circle.
- s/r ≈ 4 : this contour might be a straight line crossing the center of external circle.
- s/r becomes larger : this contour might be a curly, spiral line or multi-hollowed shape.
2. Ratio between area(A) and area of external circle(A’):
- A/A’ larger : the contour is close to circle or concentrated net structure.
- A/A’ smaller : the contour is more close to a slender line or loose net structure.
Remove and fill-in hair area
After getting mask the original image, I removed the noises from origin image, then used the inpaint function to fill-in the hair(noise) area, and recovered the image without hair.
Before in-painting, to make sure all noises including the edges of noise are all included, I first dilate the mask to ensure edges won’t become the sample for in-painting. I used the inpaint function in opencv to recover images. This function is designed based Telea’s FMM inpaint method.

The main principle of Telea’s FMM is in Fig 1. and below formulas. Every recovery work starts from a point p in the set of boundary. First, pick the p that has the largest sampling area(the area with radius epsilon.) The final value of point p is the sum of every sampling points with weighting w(p, q).

The weighting is based on three geometric factors: direction relationship, distance relationship, and level relationship. Direction relationship is calculating the distance between normal vector and sampling point q: if q is close to normal vector, it gets higher weight; vice versa. Distance relationship is depends on the distance between p and q: if they’re closer, it gains higher weight, vice versa. Last but not least, level relationship is based on the contour: if q is closer to contour, it earns higher weight; vise versa.